Welcome
The Landis Lab studies the evolution of biodiversity: how has life radiated throughout and adapted to an ever-changing world? We study evolution by developing probabilistic models, writing open source and community-minded software, and analysing simulated and empirical data.
Biological research interests for the Landis Lab include phylogenetic inference, divergence time estimation, historical biogeography, the evolution of ecological interactions, phenotypic evolution, and pathogen and parasite spread.
Our methodological interests include phylogenetic model design, stochastic processes, Bayesian inference, deep learning, and probabilistic programming.
Interested in joining? We’re always looking for creative thinkers who study evolution! We have funded positions available for postdocs and PhD students. Visit the People page for more information.
Curious to learn more? Visit the Research and Papers pages for more materials.
Lab info
Michael Landis
Assistant Professor
Department of Biology
Department of Computer Science & Engineering (courtesy appointment)
Rebstock 210, Danforth Campus
Washington Unversity in St. Louis
News
Mar 2025
- Welcome to Gabriel Santos Garcia! Gabriel is visiting us as a PhD student from University of Sao Paolo in Brazil for one year to study bat phylogenetics, biogeography and ecology. Gabriel is supported through a FAPESP BEPE scholarship.
Feb 2025
- Welcome to Anna Nagel! Anna is an expert in Bayesian modeling, popoulation genetics, and phylogenetics, and recently joined our group as a postdoc.
Oct 2024
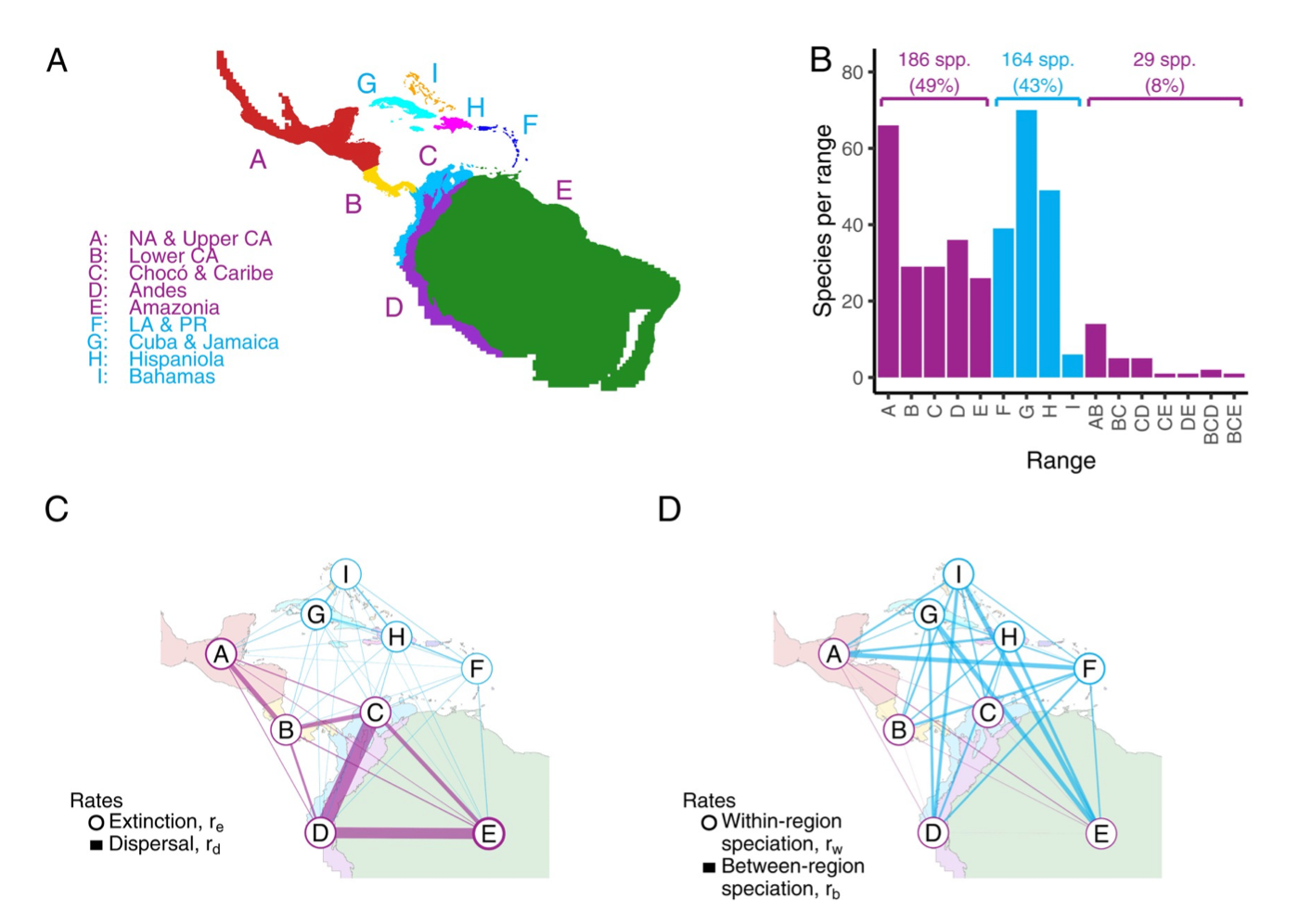
- Sarah Swiston’s paper for testing for relationships among multiple regional features and biogeographic rates was accepted in Systematic Biology. Congrats to Sarah!
Aug 2024
- Interested in simulating phylogenies? Fàbio Mendes’ new paper introducing PhyloJunction is now in advance access at Systematic Biology (link). Give it a try!
- We just launched Year 2 of the Society of Systematic Biologists’ Mentorship Program. Thanks to the new cohort of volunteers for participating and to SSB for the continued support!
Jul 2024
- New work led by Albert Soewongsono for transforming SSE models into diffusion approximations is now published in Bulletin of Mathematical Biology.
- PhD students Sarah and Sean presented their research on feature-informed GeoSSE models at the Society for the Study of Amphibians and Reptiles in Ann Arbor, Michigan. 🐸
- Check out our new package phyddle, software to explore phylogenetic models with deep learning.
Jun 2024
- We just ran our first Phylogenetic Biogeography Workshop here at Washington University [link]. New RevBayes tutorials for the FIG model are now online [link]. Thanks to everyone for participating! We’ll run a second workshop soon, hosted by Felipe Zapata at UCLA.
- NSF is now funding a new POSE project to help grow RevBayes [link] into a larger open source ecosystem for the phylogenetics community. Project is led by Tracy Heath and Ben Redelings.
May 2024
- Raymond Castillo has just joined the lab. Raymond is interested in developing phylogenetic models to better understand how genes, chromosomes, and genomes evolve in plants.
Apr 2024
- Grateful to receive WUSTL Here & Next Tier 3 funding to study host-vector-microbe disease transmission dynamics involving ticks. Part of a new collaboration with Solny Adalsteinsson, Jacco Boon, and Susan Flowers.
Jan 2024
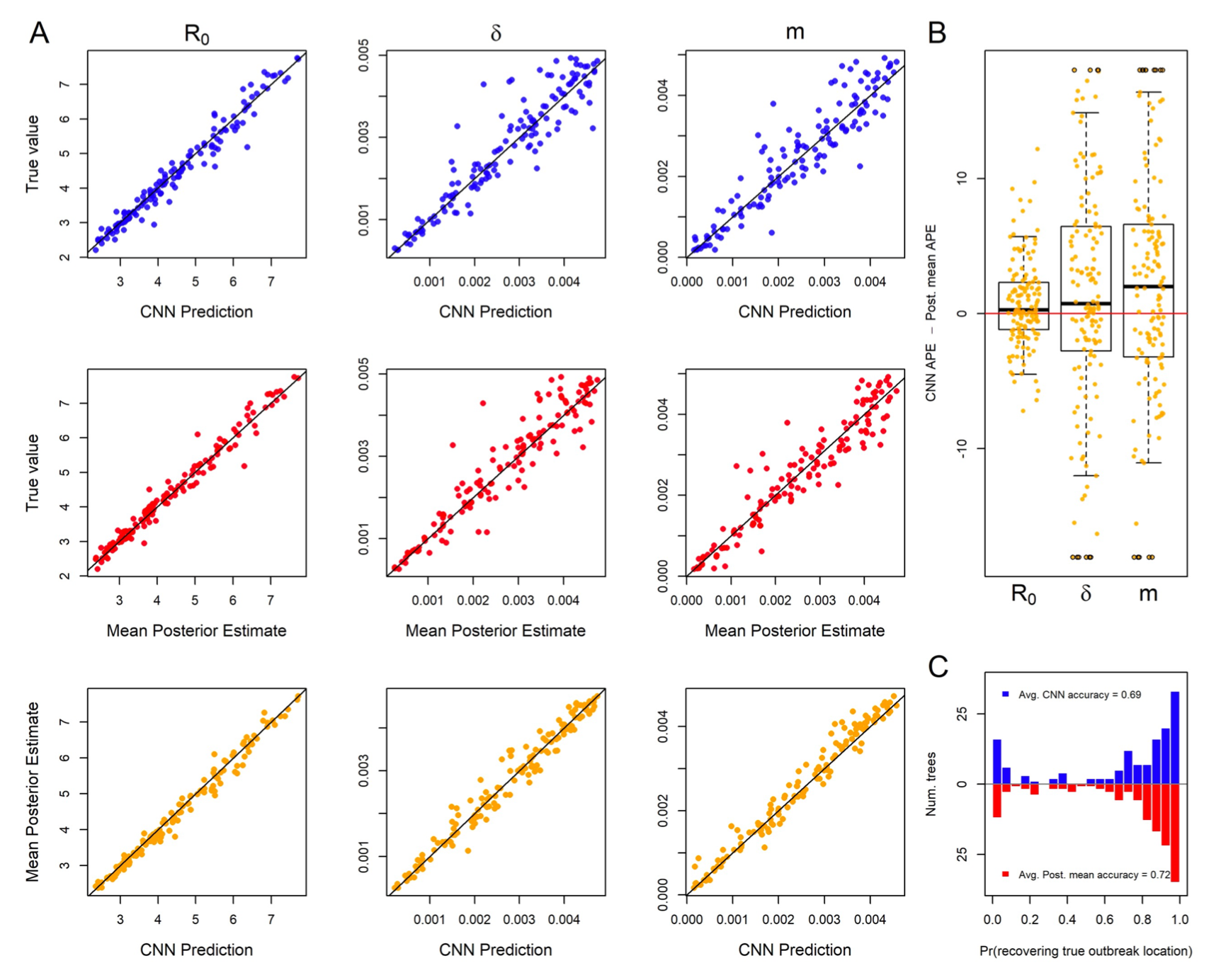
- New work led by Ammon Thompson now accepted and online in Systematic Biology. The work shows that convolutional neural networks trained for viral phylogeography perform similarly to gold-standard Bayesian methods under a range of realistic model misspecification scenarios. Some ideas captured in the Phyloseminar recording, here.
Sep 2023
- We are recruiting graduate students to study phylogenetic approaches, particularly for epidemiology using deep learning inference methods. Opportunities to join the lab through the Division of Biological & Biomedical Sciences or Computer Science & Engineering PhD programs. Email Michael with a brief description research interests and/or a recent CV to express your intent to apply.
Aug 2023
- Some good news for the lab: NIH/NSF-EEID awarded us funding to develop new Bayesian and deep learning methods to reconstruct phylogeographic spread of viruses among multiple populations and species. Collaborators are Krista Milich (Anthro), Dave Wang (Pathology & Immunology), and Innocent Rwego (Makerere University). We’ll be hiring for computational epidemiologists soon. 🦠
Jul 2023
- Our lab is hiring a postdoc to research statistical phylogenetic models and methods. We’ll design the project together, based on shared interests and types of expertise. Position is for 2+ years with $60k salary. Visit our ad on evoldir for more details.
- Also, be sure to check out our new YouTube channel with collected video recordings of research talks from our lab members. Videos are also cross-linked through papers.
Jun 2023
- Thrilled to share that Fàbio Mendes (postdoc) will be joining LSU as an assistant professor in Fall 2024! Contact him to learn about joining his new lab as grad student or postdoc. 🎉🎉
- After a year of planning, Michael has helped launch a new Mentoring Program with the Society of Systematic Biologists as a team effort with Ixchel González-Ramírez, Alonso Delgado, and Laura Kubatko. Our pilot program formed 22 mentoring pairs to help mentor young systematists, worldwide.
May 2023
- New perspective piece with Nate Upham in Science on how diverse genomic sequencing among mammal species teaches us new lessons about the evolutionary history of our clade (link).
Apr 2023
- Sean McHugh just passed his qualifying exam on ecological niche evolution. Congrats, Sean!! 🥳
- Albert Soewongsono has just joined our lab as a postdoctoral researcher to study statistical models of lineage diversification. Welcome, Albert!
Mar 2023
- We just refreshed the lab website’s Research page to now feature recent projects and papers, and the People page to now feature a new Expectations & Values document, written collaboratively by our group.
Jan 2023
- Added new websites dedicated to our NSF-funded Hawaiian plant radiation project (link) and our ITF-funded Ugandan viral phylogeography project (link).
Nov 2022
- Thanks to the WUSTL Incubator for Transdisciplinary Futures for funding our new project on viral phylogeography. In collaboration with Krista Milich (Anthropology) and Dave Wang (Pathology & Immunology).
Oct 2022
- New paper in Annual Reviews on modeling the evolution of intimate ecological interactions among species using cophylogenetic methods. Paper available here.
Sep 2022
- Sean McHugh has officially joined our group as a new PhD student. Sean is interested in how ecological traits evolve among phylogenetic lineages. Welcome, Sean!
Jul 2022
- We’re hiring a 3yr postdoc to collaborate on phylogenetic models! Details for applying are here. Feel free to email me with any questions.
May 2022
- I wrote a short article on inferring where anole ranges tend to split for Anole Annals, based on our recent paper on modeling historical bigoeography.
Apr 2022
- Two cheers for Sarah Swiston for passing her qualifying exam and for being awarded the NSF Graduate Research Fellowship! 🎉🎉
Mar 2022
- New study now published in PNAS, modeling how regional features of geography (e.g. oceans, distances) influence speciation, extinction, and dispersal rates. Read more here and here.
Oct 2021
- With the re-opening of the lab this Fall, our group welcomes three new members – Dr. Fábio Mendes (postdoc), Sarah Swiston (PhD student), and Mihir Shah (undergrad) – and bids farewell to two others – Dr. Mariana Braga (postdoc) and Ernie Ramos (undergrad).
Jul 2021
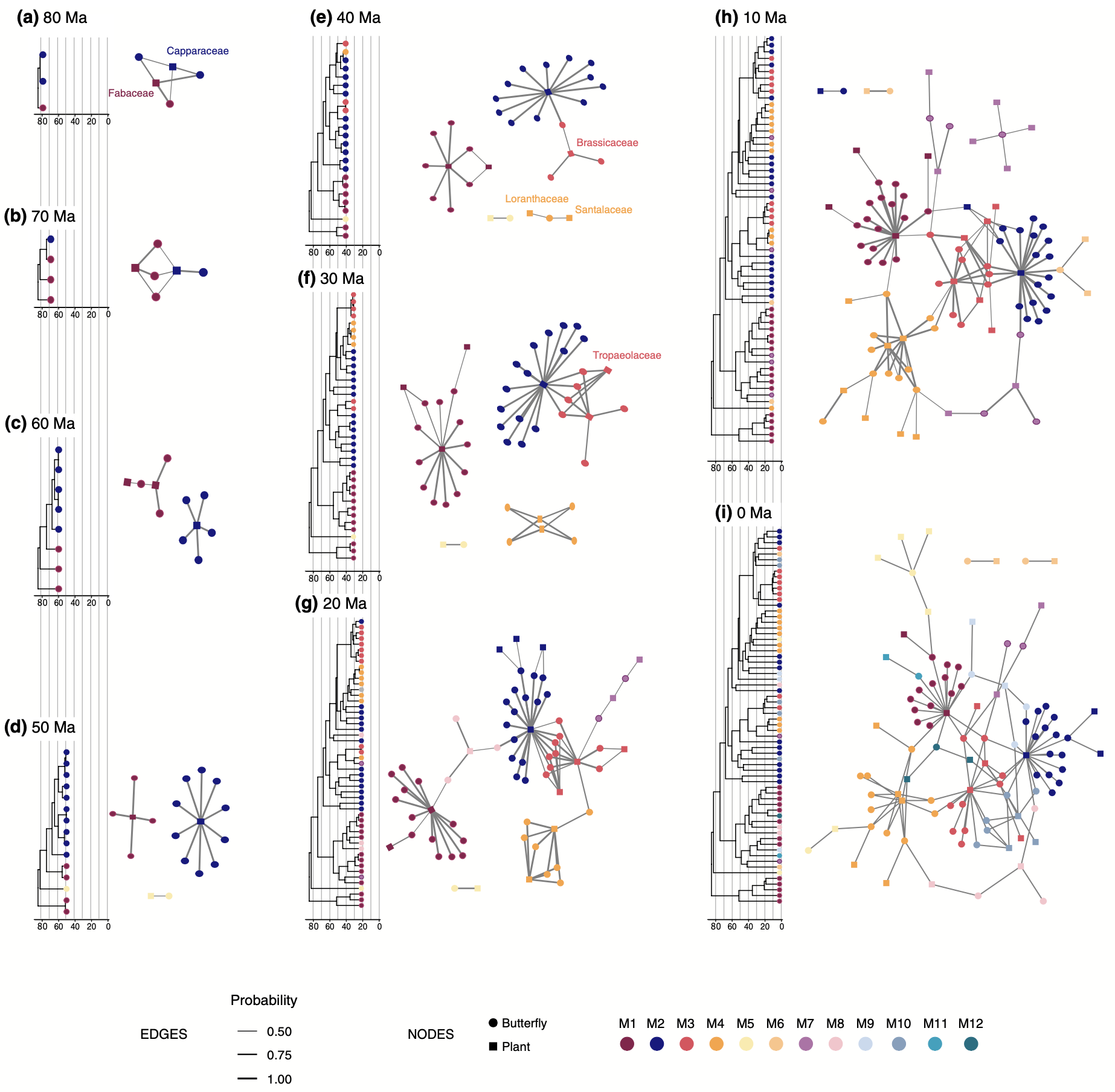
- We have a new paper out in Ecology Letters, introducing a new phylogenetic method to reconstruct how ecological networks (e.g. between plant and butterfly species) evolved over deep time scales. Read more here!
Jun 2021
- Our very own Dr. Mariana Braga won the Publisher’s Award from Systematic Biology for her paper entitled “Bayesian inference of ancestral host–parasite interactions under a phylogenetic model of host repertoire evolution”. Nicely done, Mariana!
Apr 2021
- We’re excited to welcome several new people who recently joined the lab. Dr. Ammon Thompson is a visiting scholar from the National Geospatial Intelligence Agency who is working with us to develop phylogenetic methods to predict epidemiological outcomes.
- We’re also fortunate to have two new undergraduate researchers: Ernie Ramos is a math major interested in the relationship between ecology, habitat, and evolution; and Walker Sexton is a biology major interested in evolution, systematics, and paleogenomics.
Jan 2021
- We’re hiring for a postdoc to study computational phylogenetics and biogeography. Application reviews begin 01 Feb 2021. Apply here!
Jan 2021
- Our Viburnum work featured on the cover of this year’s first issue of Systematic Biology!
Nov 2020
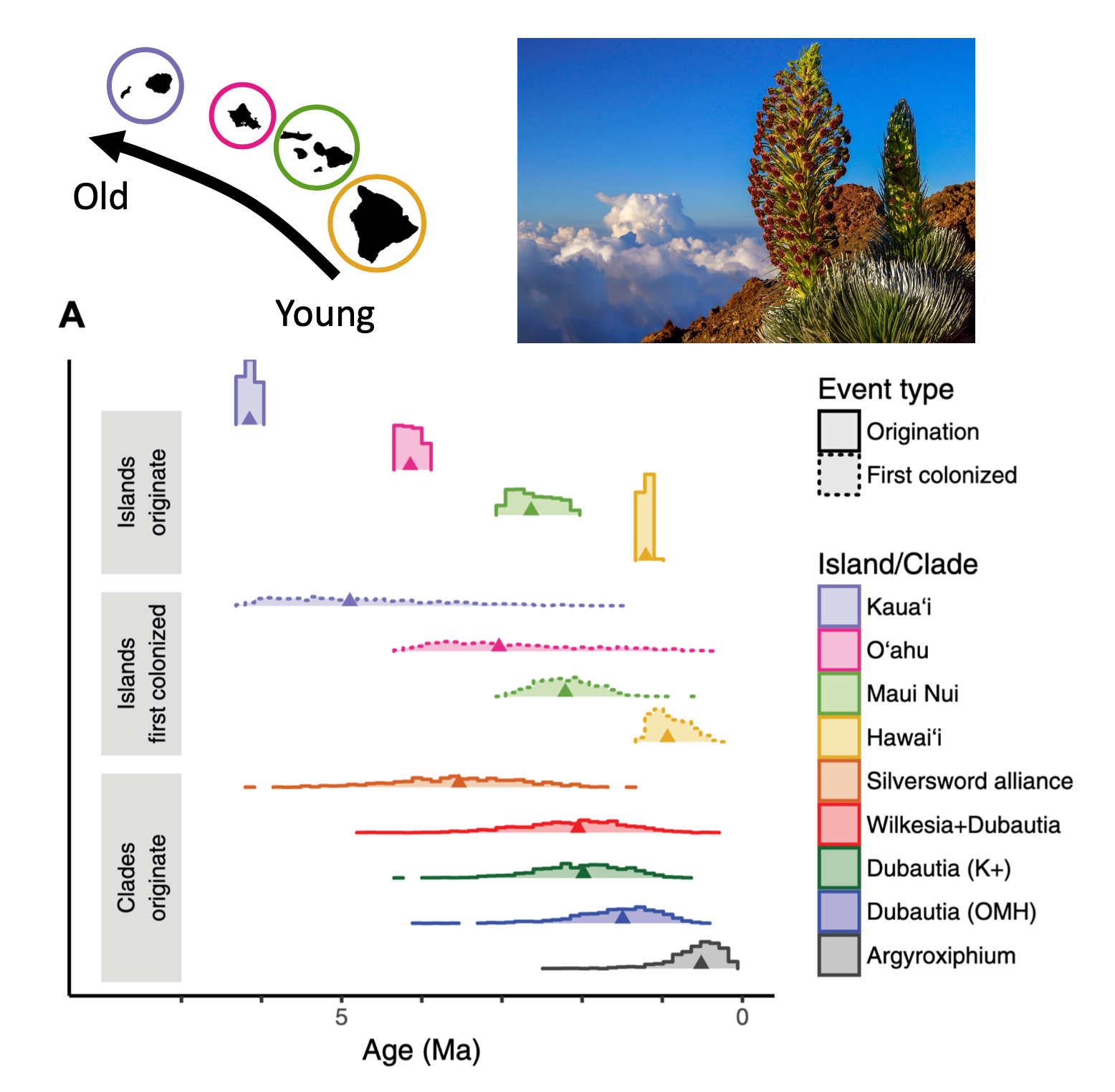
- The NSF funded our grant to model how plant lineages radiated throughout the Hawaiian archipelago! Work is in collaboration with Felipe Zapata (UCLA), Nina Ronsted (NTBG Hawaii), Warren Wagner (Smithsonian), Bruce Baldwin (UC Berkeley), and Will Freyman (23andMe).
May 2020
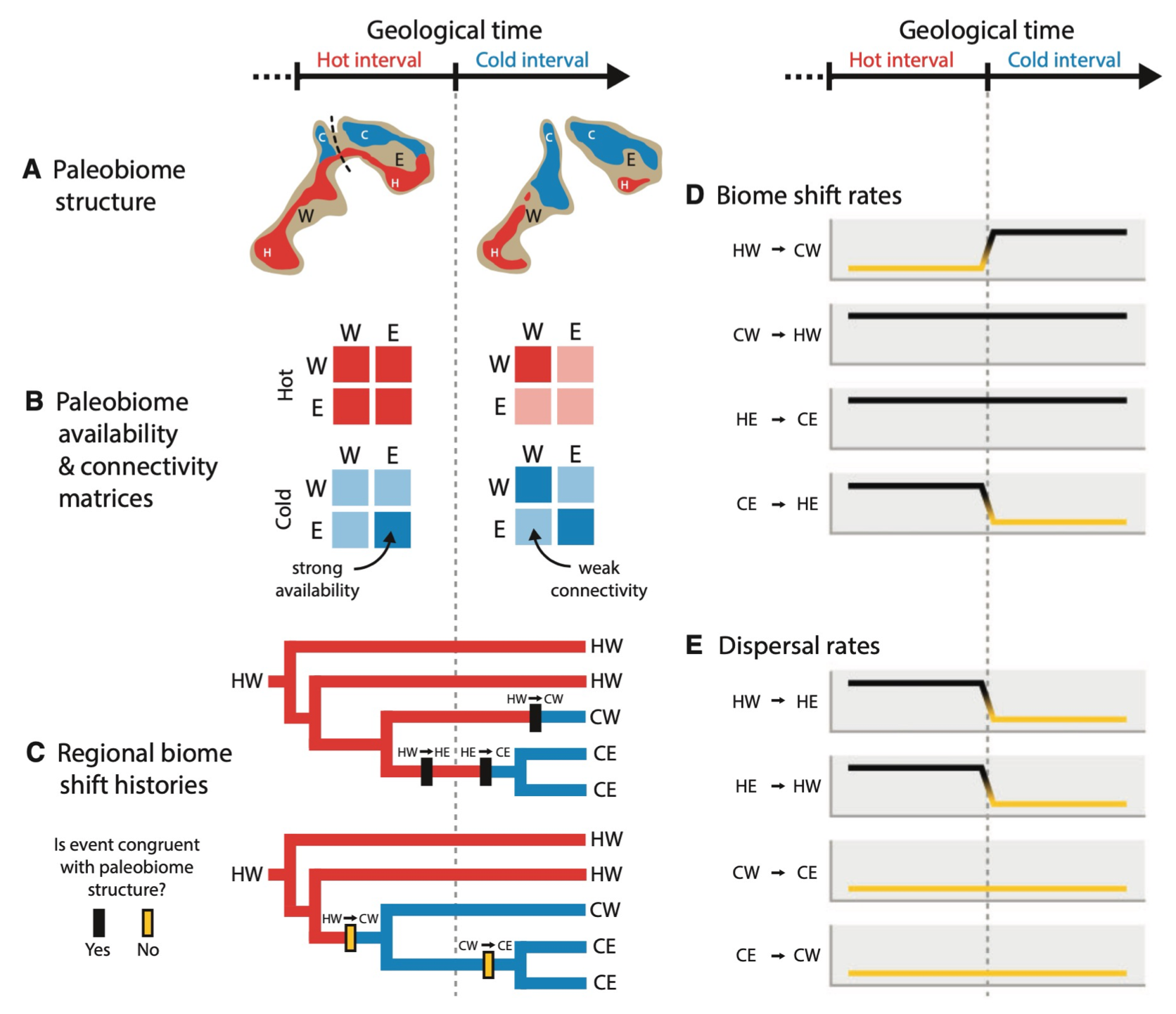
- A second new paper in Systematic Biology by Michael and Yale colleagues was accepted, which introduces an inference framework to model how lineages shift among paleobiomes, also investigating the evolution of Viburnum.
May 2020
- Our group was highlighted by the Society for the Study of Evolution in their monthly New Faculty Profiles. Read the interview here.
Apr 2020
- Michael and collaborators from Yale have a new paper in Systematic Biology that introduces a phylogenetic method to jointly infer species relationships, divergence times, and ancestral ranges and biomes from extant and fossil taxa, applied to the plant clade, Viburnum.
Mar 2020
- Mariana, Michael, and collaborators from Stockholm University and the Swedish Natural History Museum have published a new paper in Systematic Biology that introduces a phylogenetic model to estimate how the intimate species interactions co-evolved, with an application to host plant-butterfly co-evolution.
Mar 2020
- With collaborators in Wash U School of Medicine, Michael has a new paper published in Cell Host & Microbe, investigating the evolutionary origin of an insertion sequence that confers resistances to alphavirus infections, such as Chikungunya, found only cows & their close relatives.
Dec 2019
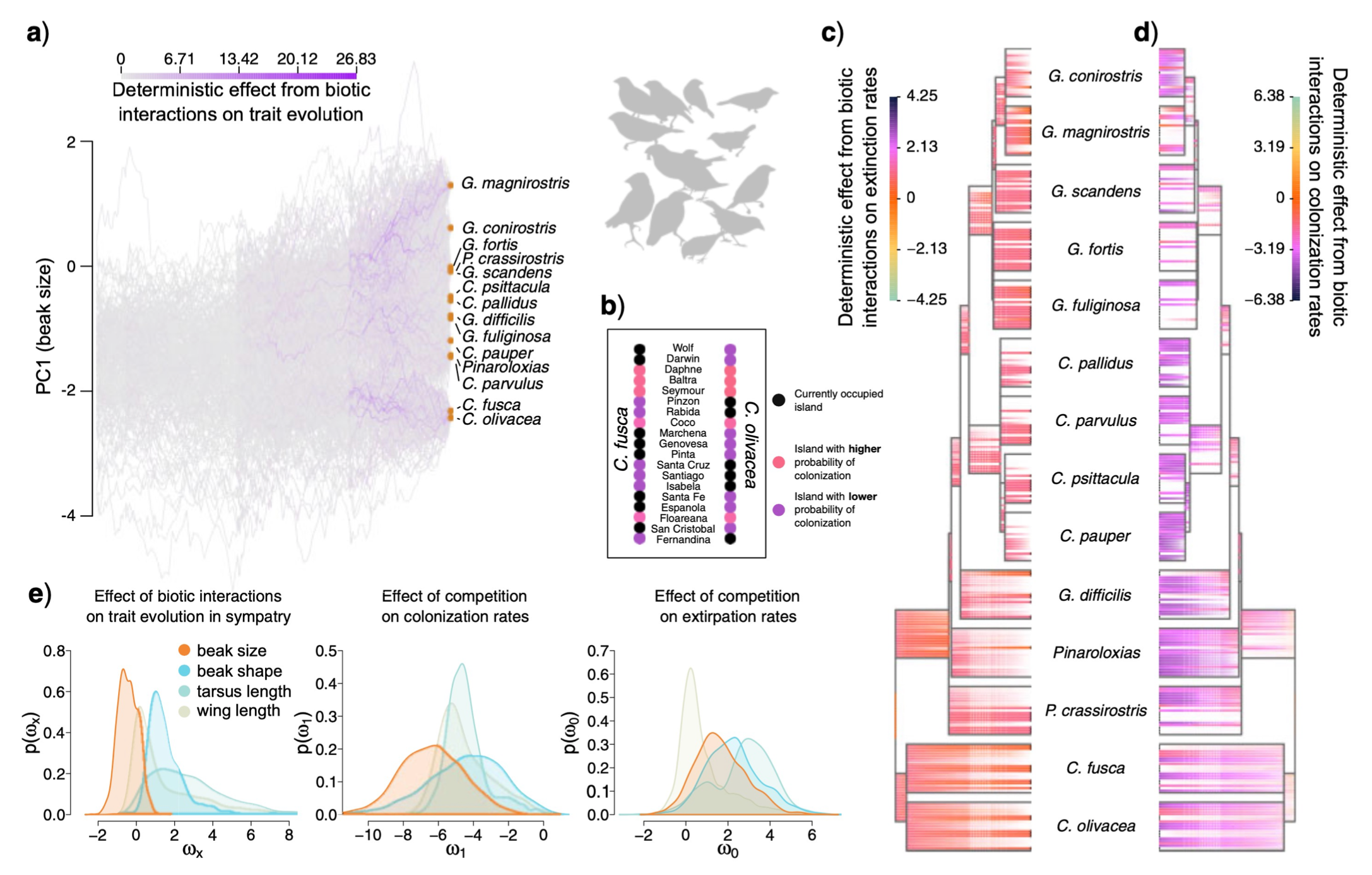
- Michael and collaborator Ignacio Quintero (ENS Paris) have a new paper that introduces a new framework for modeling the interdependent evolution of phenotypes and biogeography among co-evolving (e.g. competing) phylogenetic lineages, with an application in Darwin’s finches from the Galapagos.
Oct 2019
- Dr. Mariana Braga has joined the lab as a postdoctoral researcher. Dr. Braga recently earned her PhD from Stockholm University, where she studied the co-evolution of plant-insect interactions. Welcome!
Sep 2019
- We are actively recruiting PhD students to join us for Fall 2020! Interested applicants, please contact michael.landis@wustl.edu so we can learn more about you and your research interests, and so we can answer any questions you have about applying. Learn more about applications here.
Aug 2019
- Just returned from the Workshop on Molecular Evolution for 2019 at MBL in beautiful ctenophorous Woods Hole, MA. Our lecture and lab materials for teaching Bayesian phylogenetics using RevBayes are hosted here – resources compiled by Dr. Tracy Heath!
Aug 2018
- We are seeking researchers interested in studying evolutionary biology from a quantitative perspective.
New lab members will design statistical models, develop scientific software methods, lead projects, and publish first-author papers.
Postdoctoral and PhD student positions are to start in Summer/Fall 2019 (some flexibility).
If interested, please email a brief statement of your research interests and a copy of your CV to michael.landis@wustl.edu.
A broader invitation to apply will be published by December 1, 2018.