Research
Models of evolutionary processes help us understand how biodiversity is generated, sustained, and lost. We study evolution by developing probabilistic models, writing open source and community-minded software, and analysing simulated and empirical data.
Biological research interests for the Landis Lab include phylogenetic inference, divergence time estimation, historical biogeography, the evolution of ecological interactions, phenotypic evolution, and pathogen and parasite spread.
Our methodological interests include phylogenetic model design, stochastic processes, Bayesian inference, deep learning, and probabilistic programming.
We maintain a broad and active interest in problems involving phylogenetic modeling and inference, and develop for RevBayes, an open-source package for modeling evolutionary processes and estimating trees. Below are some examples of recent projects and studies.
Recorded talks
Visit our Youtube channel to see recorded talks and other videos, or watch through the playlist embedded below. [link]
Funded projects
We are extremely grateful to the agencies and instutitions for funding these projects:
- Modeling the Origin and Evolution of Hawaiian Plants. Funded by NSF-DEB. Collaborators: Felipe Zapata, Nina Ronsted, Warren Wagner, Fabio Mendes, Isaac Lichter-Marck, Sarah Swiston. [link]
- Phylogenetic modeling of viral transmission dynamics at the human-wildlife interface in Uganda Funded by WUSTL-ITF. Collaborators: Krita Milich, Dave Wang, Innocent Rwengo. [link]
Deep learning and viral phylogeography
To what extent do viruses spread at different rates within and between populations?
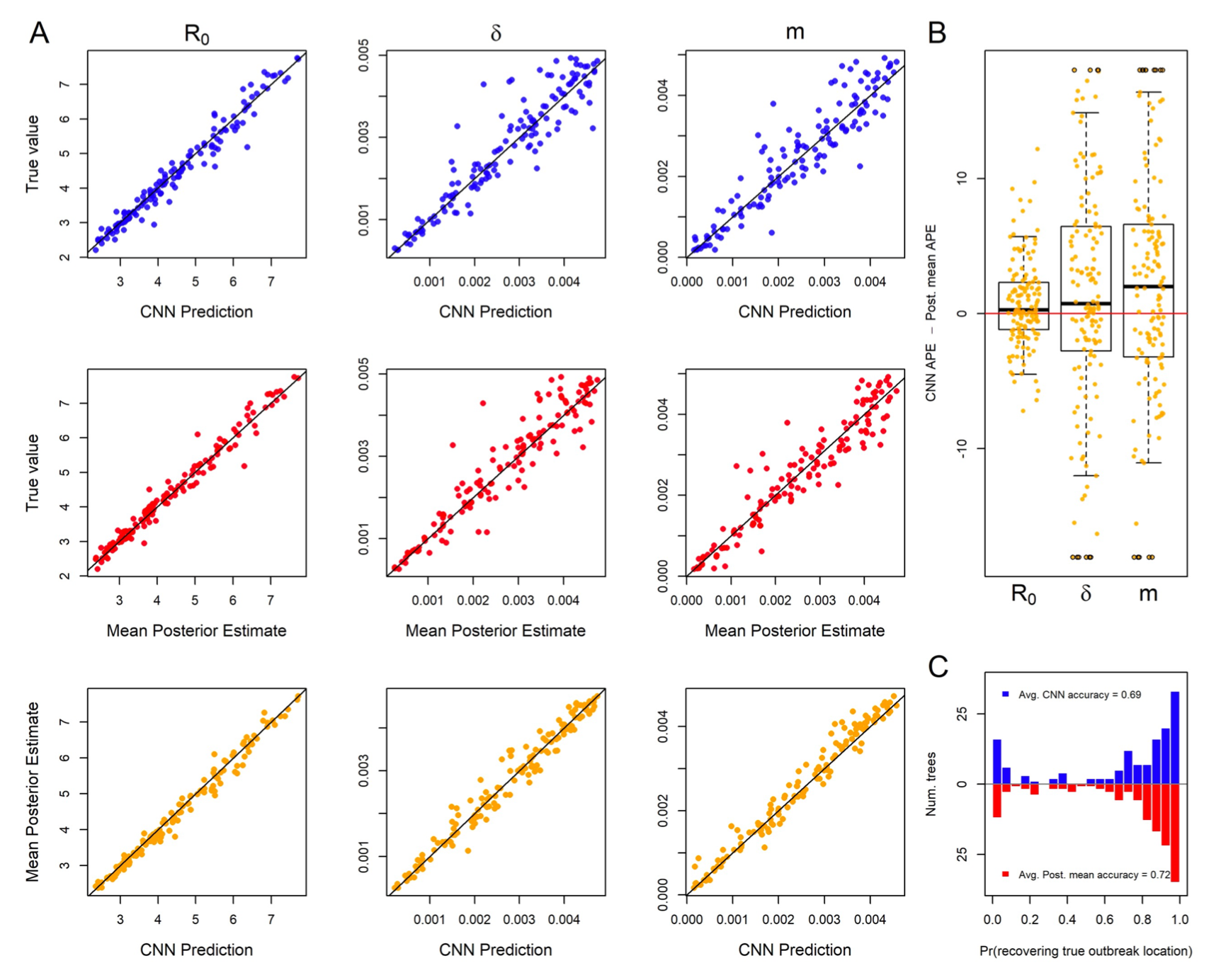 A Thompson, B Liebeskind, EJ Scully, MJ Landis. 2023. Deep learning approaches to viral phylogeography are fast and as robust as likelihood methods to model misspecification. bioRxiv 2023.02.08.527714. [paper]
A Thompson, B Liebeskind, EJ Scully, MJ Landis. 2023. Deep learning approaches to viral phylogeography are fast and as robust as likelihood methods to model misspecification. bioRxiv 2023.02.08.527714. [paper]
Biogeographic diversification
How do regional features influence rates of speciation, extinction, and dispersal?
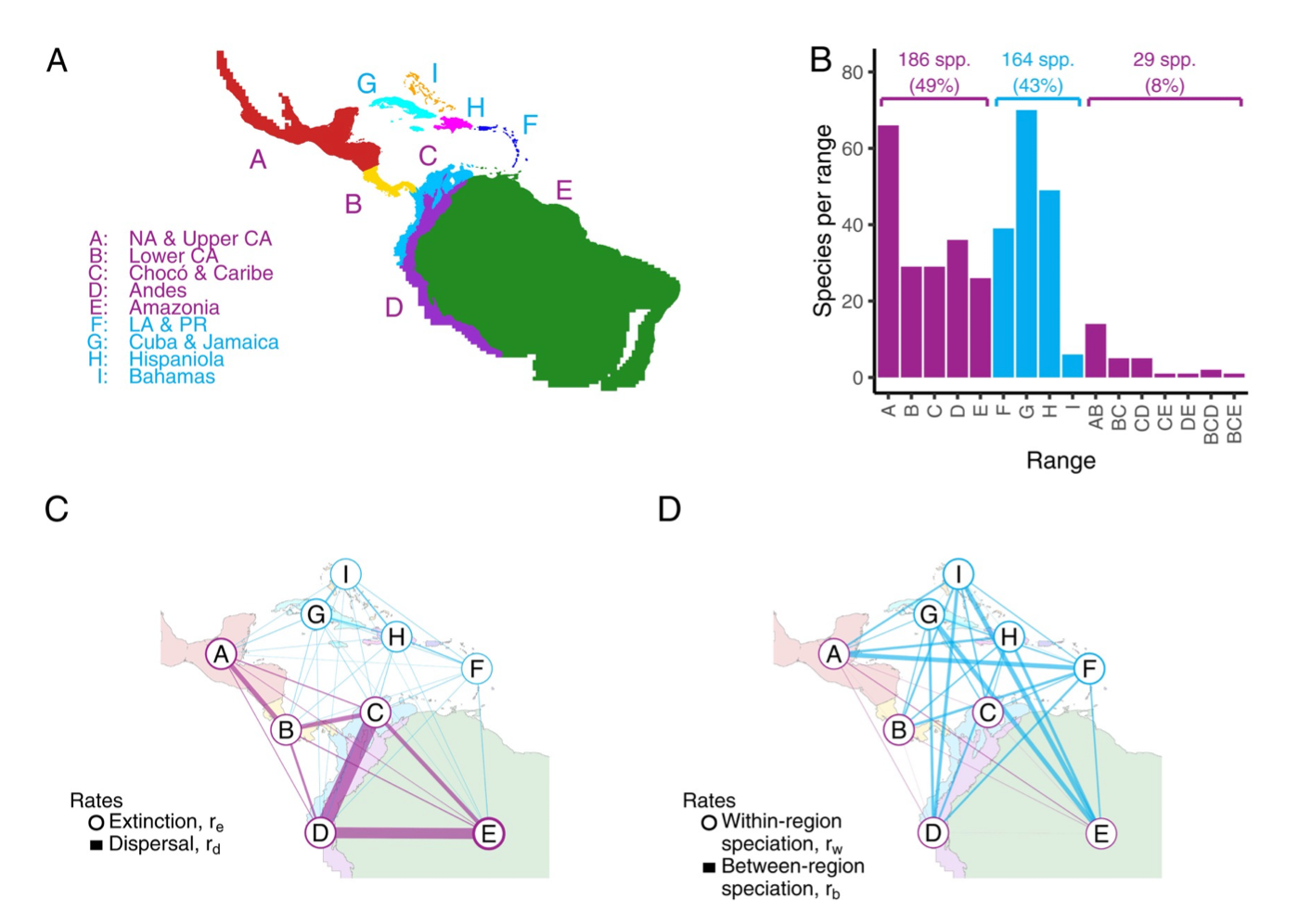 MJ Landis, I Quintero, MM Muñoz, F Zapata, MJ Donoghue. 2022. Phylogenetic inference of where species spread or split
across barriers. Proceedings of the National Academy of Sciences 119: e2116948119. [paper]
MJ Landis, I Quintero, MM Muñoz, F Zapata, MJ Donoghue. 2022. Phylogenetic inference of where species spread or split
across barriers. Proceedings of the National Academy of Sciences 119: e2116948119. [paper]
Phylogenetic biome shifts
How does geographical opportunity limit when and where species adapt to new biomes?
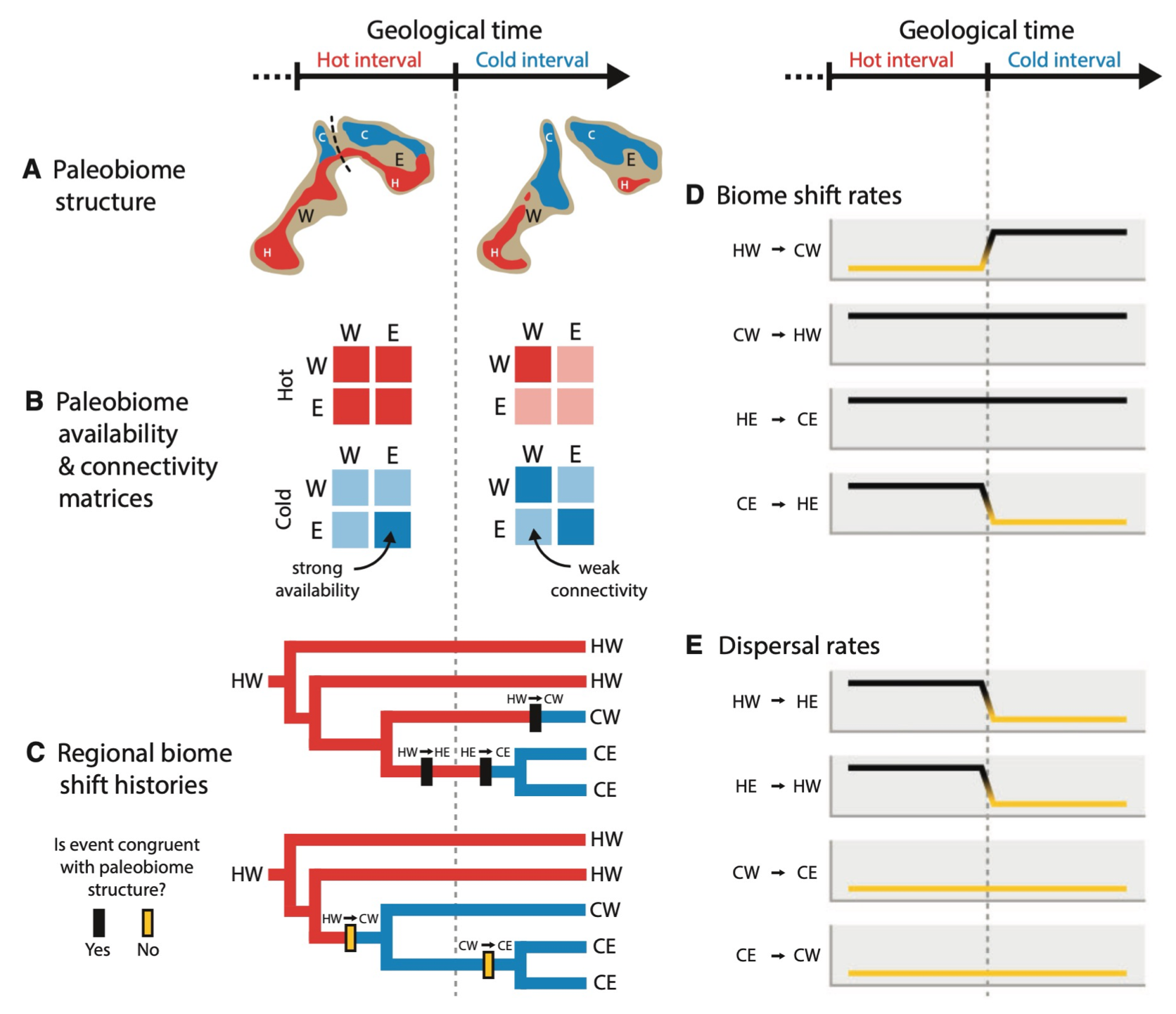 MJ Landis, EJ Edwards, MJ Donoghue. 2021. Modeling phylogenetic biome shifts on a planet with a past. Systematic Biology 70: 86-107. [paper]
MJ Landis, EJ Edwards, MJ Donoghue. 2021. Modeling phylogenetic biome shifts on a planet with a past. Systematic Biology 70: 86-107. [paper]
Evolution of ecological networks
How have symbiotic species gained and lost the ability to interact over time?
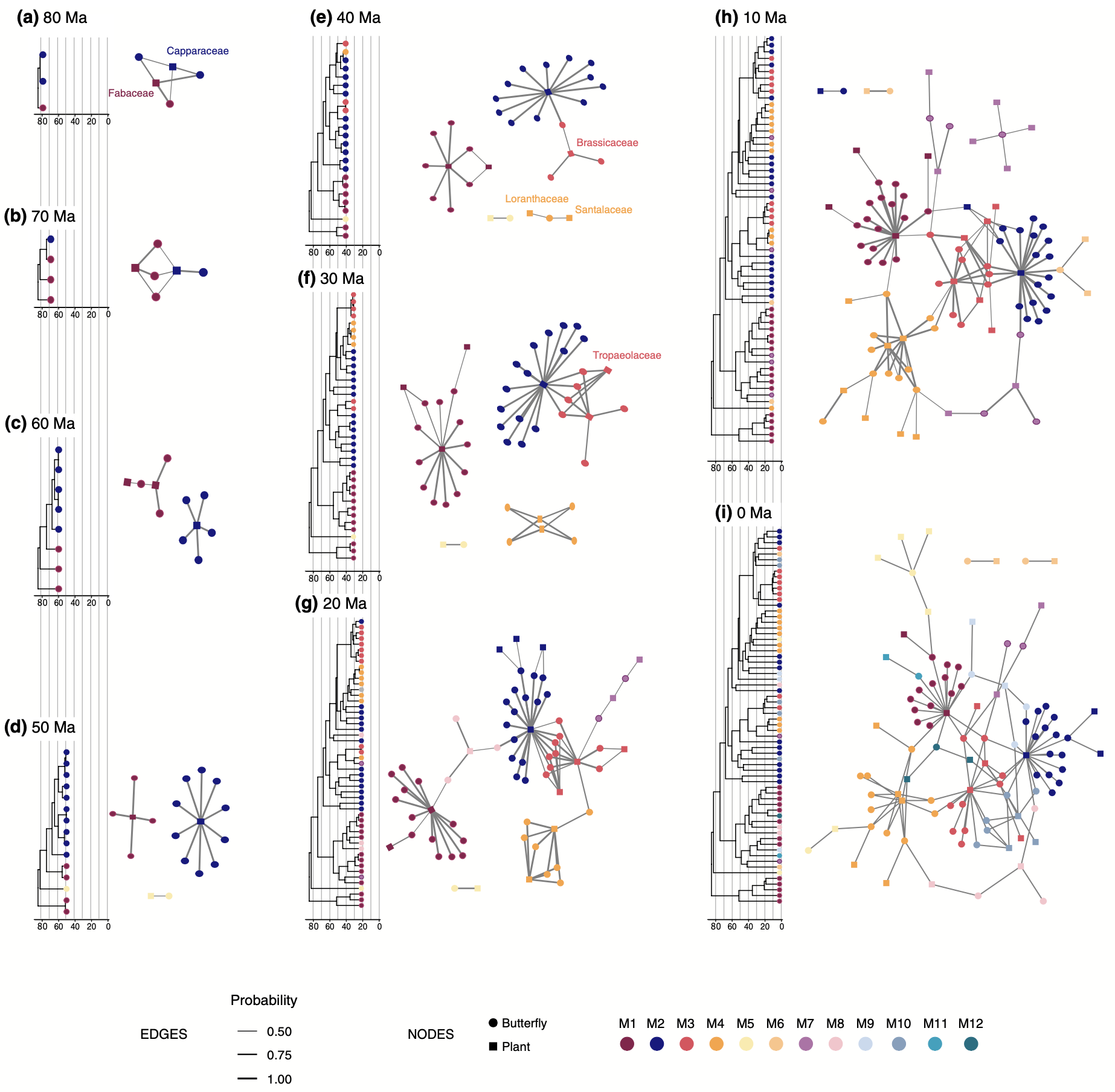 MP Braga, N Janz, S Nylin, F Ronquist, MJ Landis. 2021. Phylogenetic reconstruction of ancestral ecological networks through time for pierid butterflies and their host plants. Ecology Letters 24: 2134-2145. [paper]
MP Braga, N Janz, S Nylin, F Ronquist, MJ Landis. 2021. Phylogenetic reconstruction of ancestral ecological networks through time for pierid butterflies and their host plants. Ecology Letters 24: 2134-2145. [paper]
Evolution under competition
How do competing species evolve as a community to partition traits and space to access resources?
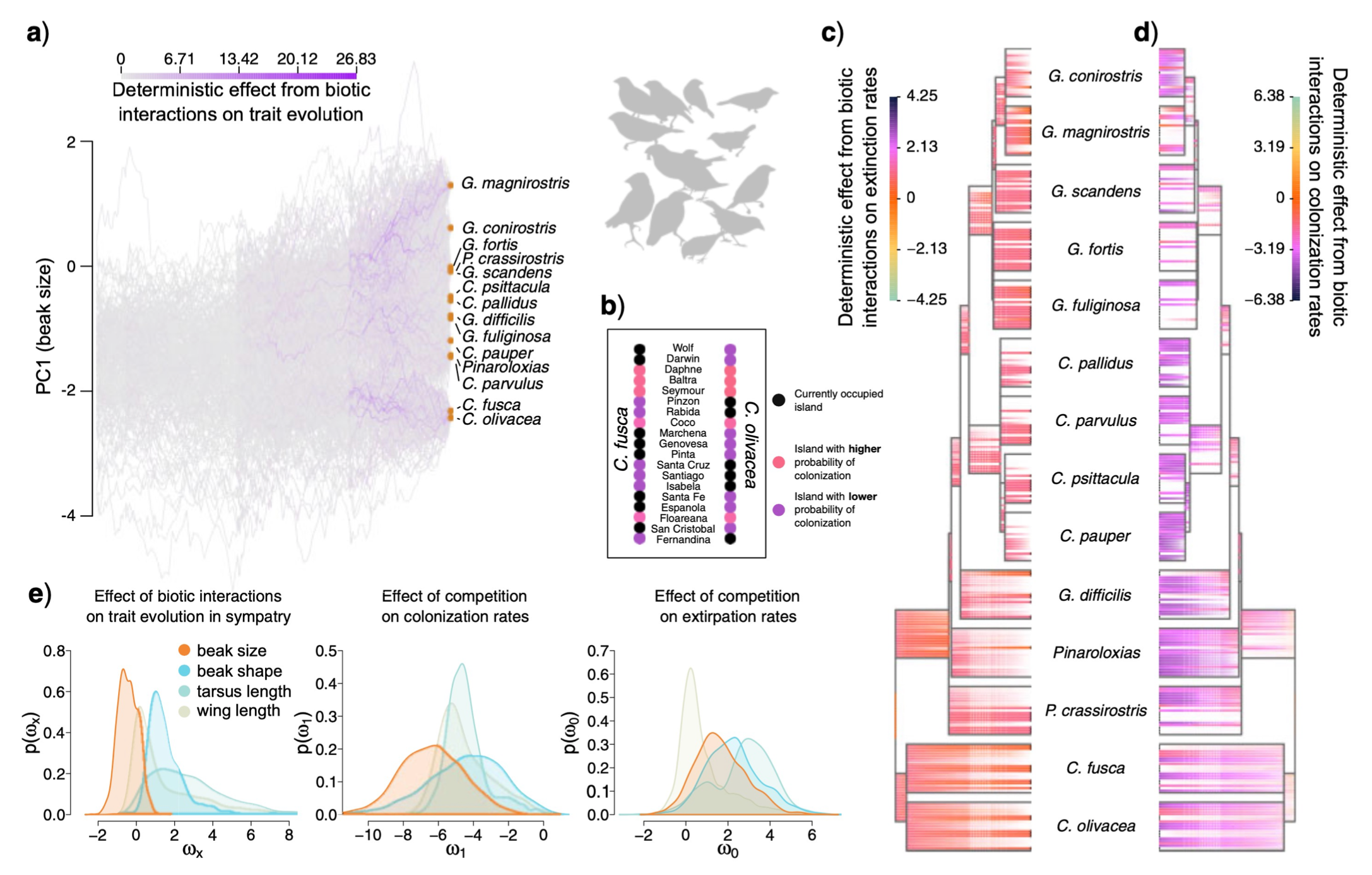 I Quintero, MJ Landis 2020. Interdependent phenotypic and biogeographic evolution driven by biotic interactions. Systematic Biology 69: 739-755. [paper]
I Quintero, MJ Landis 2020. Interdependent phenotypic and biogeographic evolution driven by biotic interactions. Systematic Biology 69: 739-755. [paper]
Biogeographic dating
How can we estimate the ages of species from genetic, biogeographic, and paleogeographic evidence?
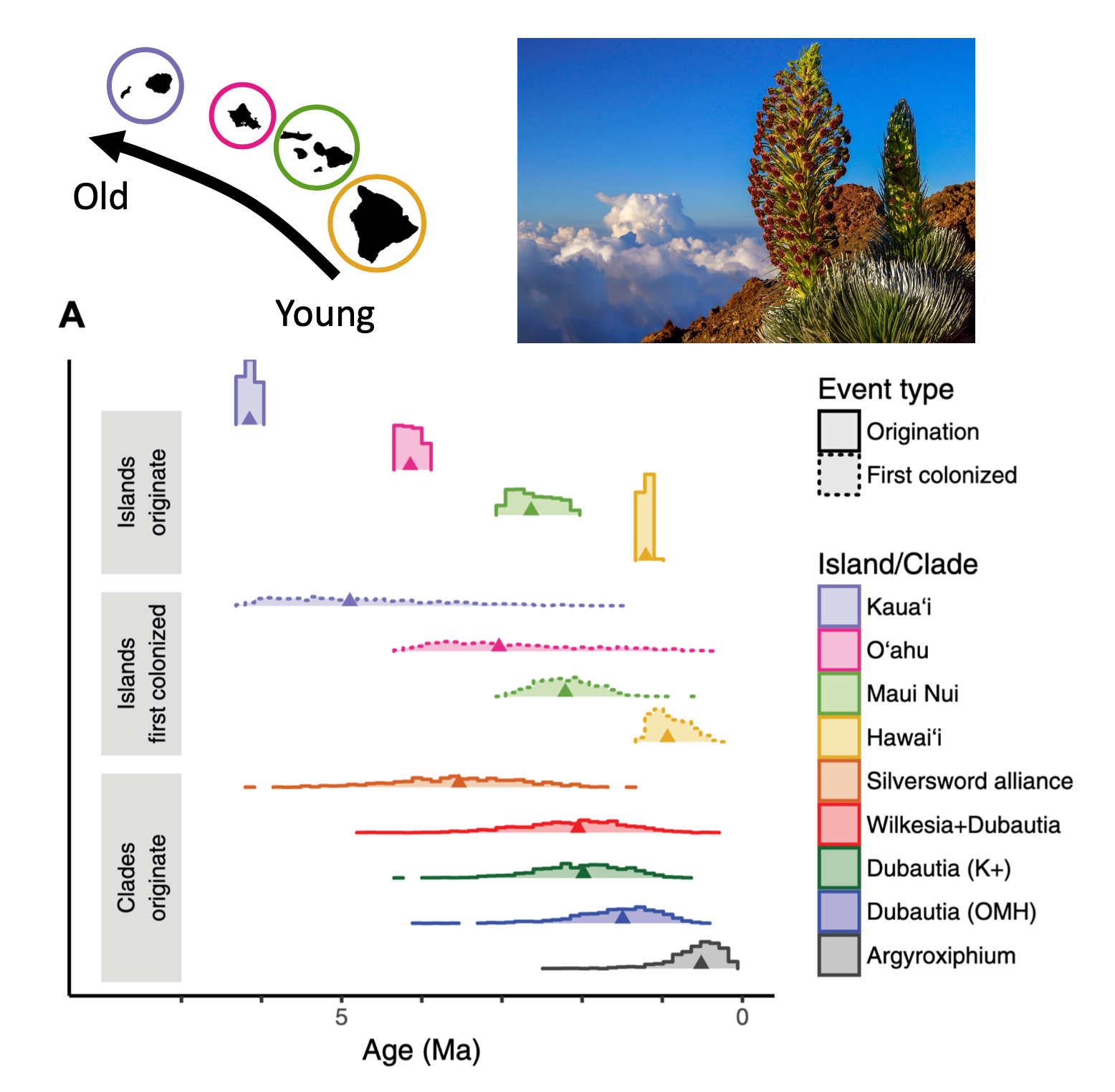 MJ Landis, WA Freyman, BG Baldwin. 2018. Retracing the silversword radiation despite phylogenetic, biogeographic, and paleogeographic uncertainty. Evolution, 72: 2343-2359. [paper]
MJ Landis, WA Freyman, BG Baldwin. 2018. Retracing the silversword radiation despite phylogenetic, biogeographic, and paleogeographic uncertainty. Evolution, 72: 2343-2359. [paper]
Tempo & mode of phenotypic evolution
Does evolution accumulate change through many small steps or through rare but large pulses?
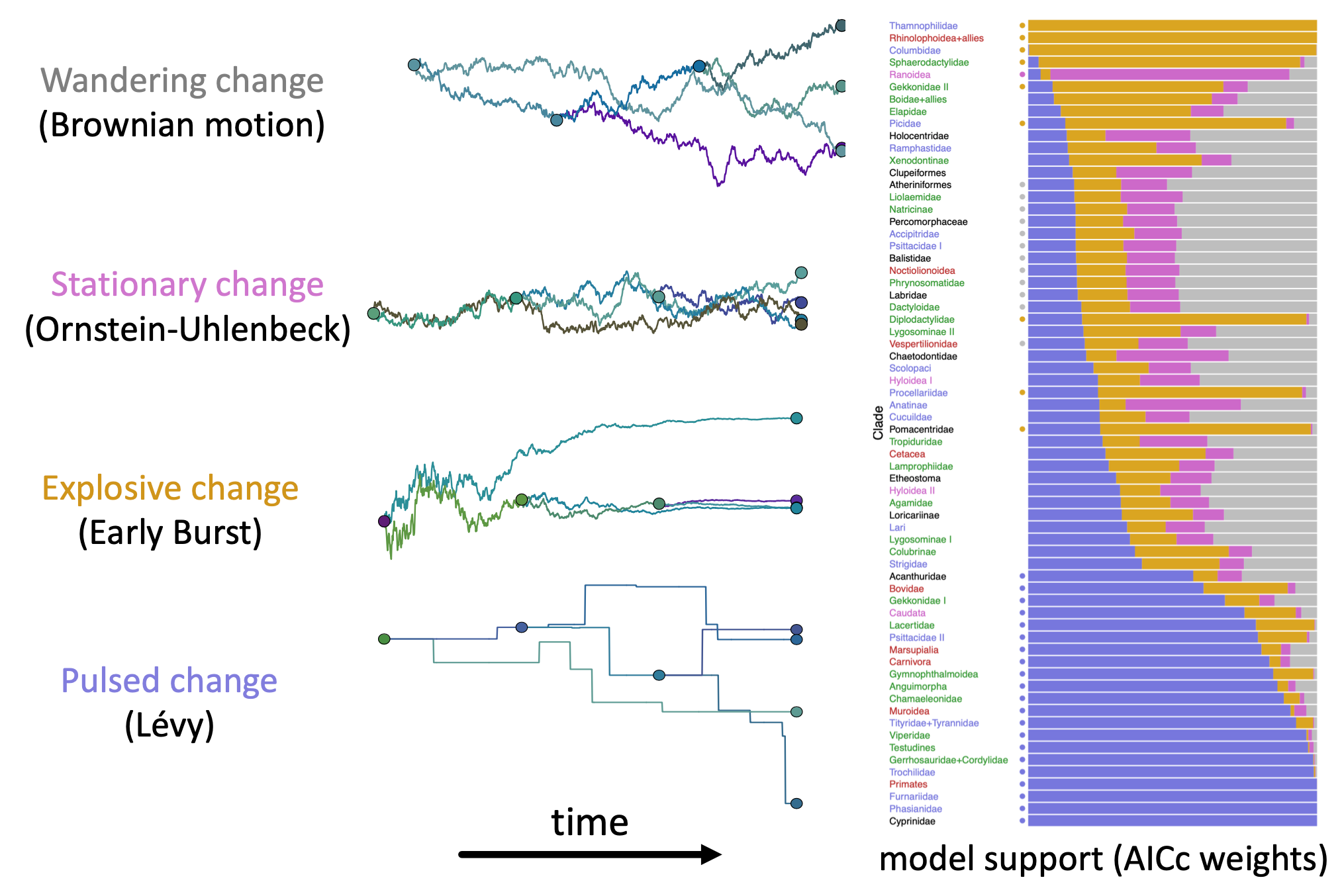 MJ Landis, JG Schraiber. 2017. Pulsed evolution shaped modern vertebrate body sizes. Proceedings of the National Academy of Sciences, 114: 13224-13229. [paper]
MJ Landis, JG Schraiber. 2017. Pulsed evolution shaped modern vertebrate body sizes. Proceedings of the National Academy of Sciences, 114: 13224-13229. [paper]
Phylogenetic graphical models
How can you build complex phylogenetic models from simpler components?
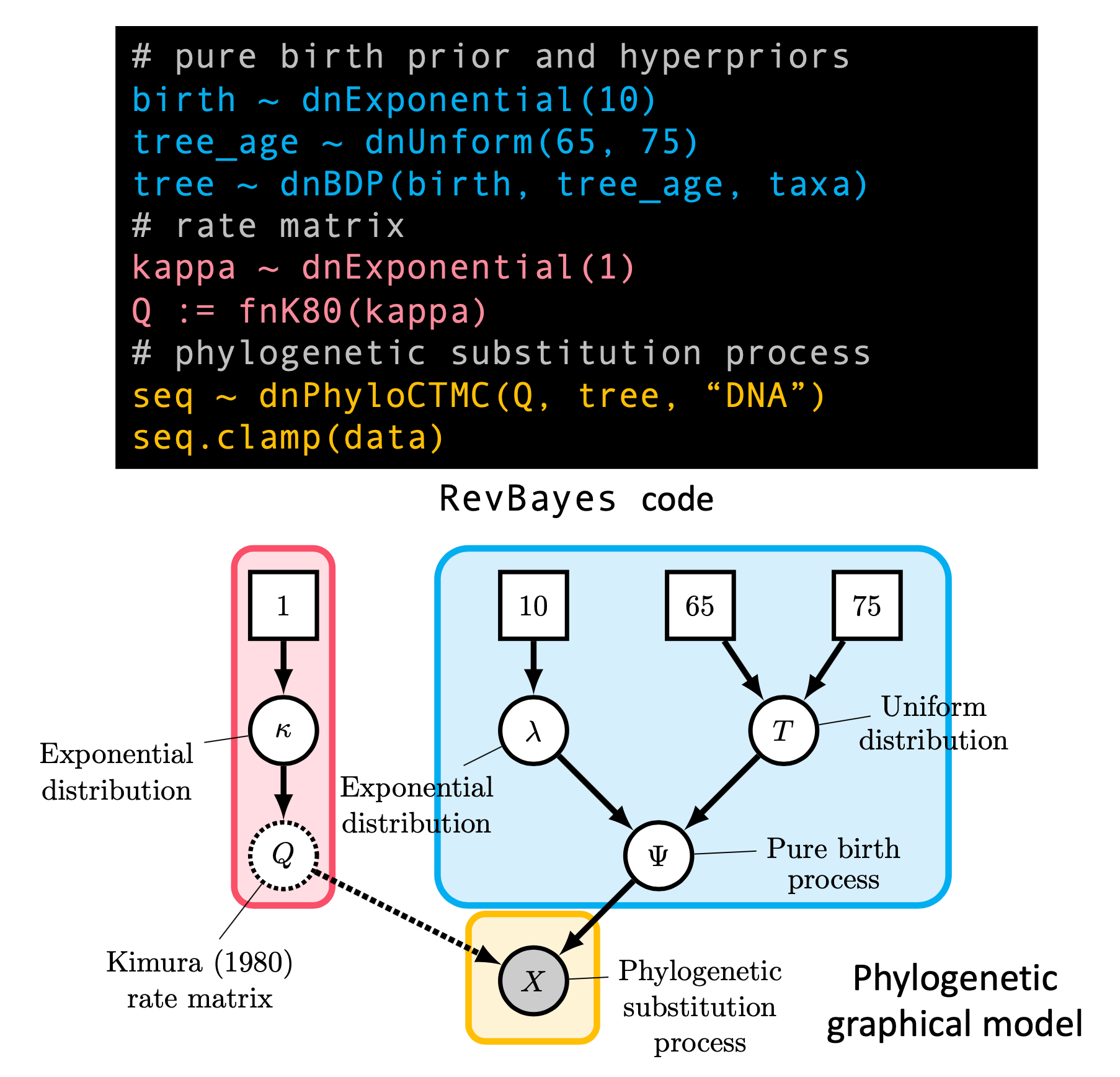 S Höhna, MJ Landis, TA Heath, B Boussau, N Lartillot, BR Moore, JP Huelsenbeck, F Ronquist. 2016. RevBayes: Bayesian Phylogenetic Inference Using Graphical Models and an Interactive Model-Specification Language. Systematic Biology, 65: 726-736. [paper]
S Höhna, MJ Landis, TA Heath, B Boussau, N Lartillot, BR Moore, JP Huelsenbeck, F Ronquist. 2016. RevBayes: Bayesian Phylogenetic Inference Using Graphical Models and an Interactive Model-Specification Language. Systematic Biology, 65: 726-736. [paper]